 Walid M. Abdelmoula, Ricardo J. Carreira, Reinald Shyti, Benjamin Balluff, René J. M. van Zeijl, Else A. Tolner, Boudewijn F. P. Lelieveldt, Arn M. J. M. van den Maagdenberg, Liam A. McDonnell and Jouke Dijkstra
Walid M. Abdelmoula, Ricardo J. Carreira, Reinald Shyti, Benjamin Balluff, René J. M. van Zeijl, Else A. Tolner, Boudewijn F. P. Lelieveldt, Arn M. J. M. van den Maagdenberg, Liam A. McDonnell and Jouke Dijkstra
Anal Chem. 2014 Apr 15;86(8):3947-54. Epub 2014 Apr 2.
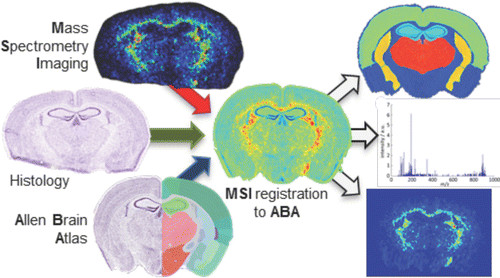
Mass spectrometry imaging holds great potential for understanding the molecular basis of neurological disease. Several key studies have demonstrated its ability to uncover disease-related biomolecular changes in rodent models of disease, even if highly localized or invisible to established histological methods. The high analytical reproducibility necessary for the biomedical application of mass spectrometry imaging means it is widely developed in mass spectrometry laboratories. However, many lack the expertise to correctly annotate the complex anatomy of brain tissue, or have the capacity to analyze the number of animals required in preclinical studies, especially considering the significant variability in sizes of brain regions. To address this issue, we have developed a pipeline to automatically map mass spectrometry imaging data sets of mouse brains to the Allen Brain Reference Atlas, which contains publically available data combining gene expression with brain anatomical locations. Our pipeline enables facile and rapid interanimal comparisons by first testing if each animal’s tissue section was sampled at a similar location and enabling the extraction of the biomolecular signatures from specific brain regions.

No responses yet