 Karolina Škrášková, Artem Khmelinskii, Walid M. Abdelmoula, Stephanie De Munter, Myriam Baes, Liam McDonnell, Jouke Dijkstra and Ron M. A. Heeren
Karolina Škrášková, Artem Khmelinskii, Walid M. Abdelmoula, Stephanie De Munter, Myriam Baes, Liam McDonnell, Jouke Dijkstra and Ron M. A. Heeren
J Am Soc Mass Spectrom. 2015 Jun;26(6):948-57. Epub 2015 Apr 28.
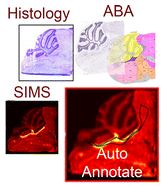
Mass spectrometry imaging (MSI) is a powerful tool for the molecular characterization of specific tissue regions. Histochemical staining provides anatomic information complementary to MSI data. The combination of both modalities has been proven to be beneficial. However, direct comparison of histology based and mass spectrometry-based molecular images can become problematic because of potential tissue damages or changes caused by different sample preparation. Curated atlases such as the Allen Brain Atlas (ABA) offer a collection of highly detailed and standardized anatomic information. Direct comparison of MSI brain data to the ABA allows for conclusions to be drawn on precise anatomic localization of the molecular signal. Here we applied secondary ion mass spectrometry imaging at high spatial resolution to study brains of knock-out mouse models with impaired peroxisomal β-oxidation. Murine models were lacking D-multifunctional protein (MFP2), which is involved in degradation of very long chain fatty acids. SIMS imaging revealed deposits of fatty acids within distinct brain regions. Manual comparison of the MSI data with the histologic stains did not allow for an unequivocal anatomic identification of the fatty acids rich regions. We further employed an automated pipeline for co-registration of the SIMS data to the ABA. The registration enabled precise anatomic annotation of the brain structures with the revealed lipid deposits. The precise anatomic localization allowed for a deeper insight into the pathology of Mfp2 deficient mouse models.

No responses yet